

Depending on the steps required by your specific application, processed data may have properties different from the original files. For instance, it may consist of a different number of channels, either because signals from more than a single recording device were merged (see Example 2) or because one or more defective channels were discarded (using ndm_extractchannels). The sampling rate may also have changed (using ndm_resample). Even the video frame rate can be altered (by the LED tracking program spots2pos, a Matlab function included in the NDManager Plugins).
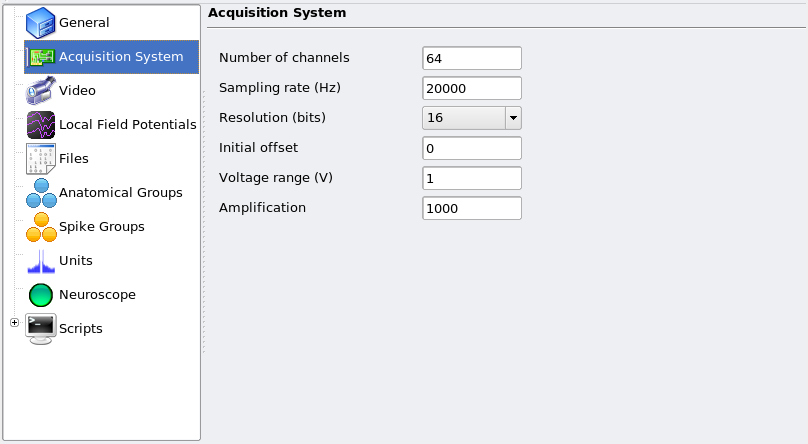
The Acquisition System page should reflect the processed data, i.e. it must describe the virtual system that would have been used to yield the data. Consider this example: data was recorded using two systems in parallel, each sampling 32 channels at 19531.25 Hz, and you wish to merge the two and resample at 20 kHz. The Acquisition System page should look like:
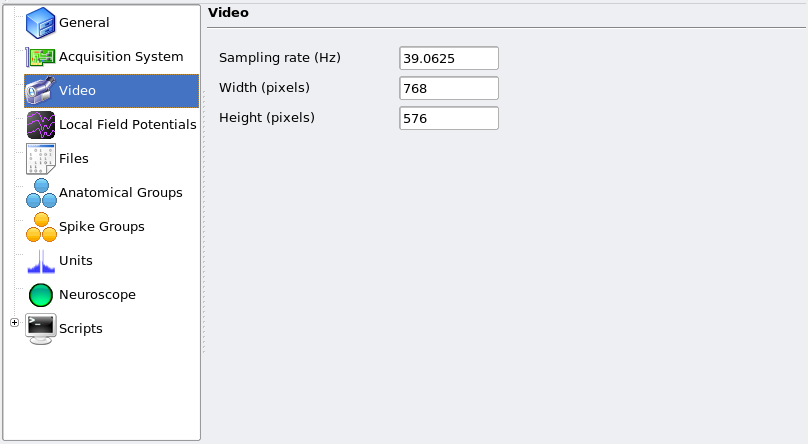
Similarly, if the video camera sampled frames at 25 Hz but tracking was resampled at 39.0625 Hz, then the Video page should look like:
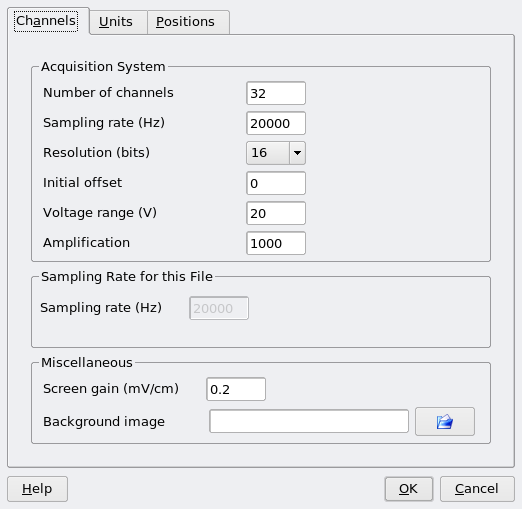
Parameter files can be created and graphically edited using NeuroScope. If you open a wideband data file (.dat file) but do not have a corresponding parameter file (.xml file), NeuroScope will present a dialog to gather minimum information and create one for you.
Also, if none of the standard template files suits your needs and you must create a template parameter file from scratch, it is a tedious process to manually create anatomical groups and spike groups (imagine manually entering all values shown in Example 1). An easier way to achieve this is to create and edit the groups using the palettes in NeuroScope.
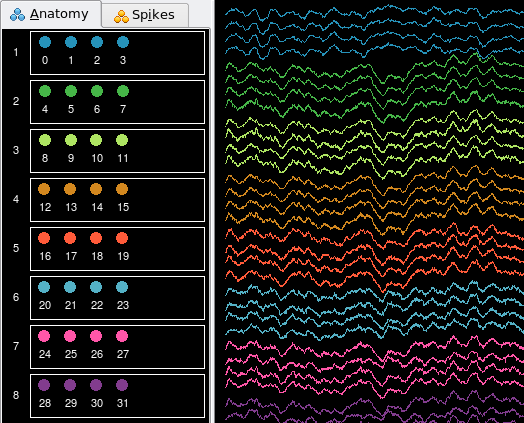
Channel colors will also be saved in the parameter file. This can be used to easily improve an existing parameter file in order to create a template parameter file for subsequent batch processing.
The drawback is that obviously this requires an existing wideband data file (.dat file), which will typically not be available until the data was processed. One solution to this chicken-and-egg problem is to process the data for one day using a hand-made template parameter file, then open the wideband data file with NeuroScope and edit the groups, then copy the parameter file as a new template.
Would you like to make a comment or contribute an update to this page?
Send feedback to the KDE Docs Team