

The example data consists of two days of recordings (July 18-19, 2011). On the first day, two sessions were run: baseline in home cage and run on a maze. On the second day, there was a third session: sleep. The recording setup used Neuralynx acquisition software (Neuralynx, Bozeman, USA). Brain signals were sampled at 32552.083 Hz from 2 tetrodes (8 channels), and video was sampled at 25 Hz at 768x576 pixels.
Initially, the data files are located in five different directories:
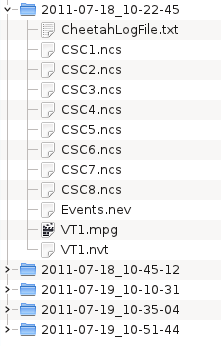
Right-click both directories corresponding to the first day, and select ->. This only works with a KDE file manager, but the same result can be achieved from a shell by typing:
ndm_prepare 2011-07-18_10-22-45 2011-07-18_10-45-12This will start a graphical dialog where you can provide the information required to rename the files:
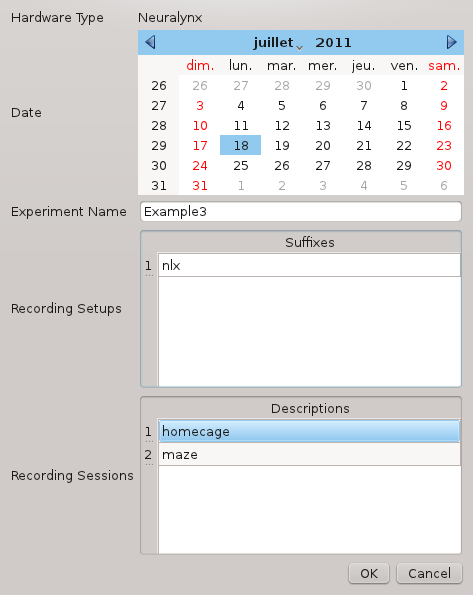
The Recording Setups entry is an arbitrary suffix that will be used to separate the original and processed files (see below). Here we choose nlx. A more advanced use case is presented in Example 2.
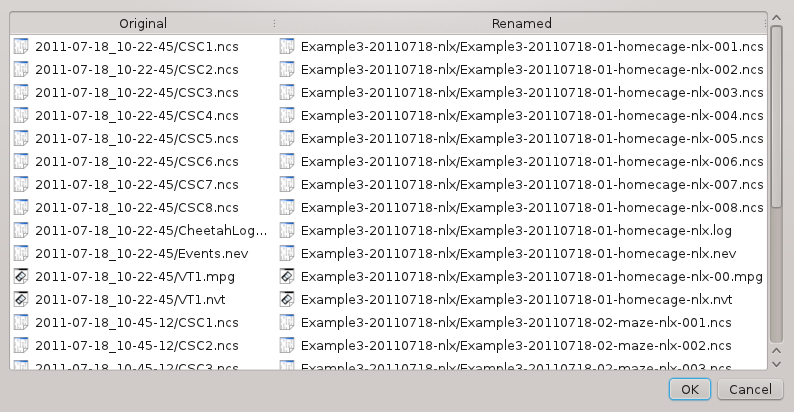
Click . In the next dialog, you can check and confirm the information you provided before the files are actually renamed:
Click . You are now presented with a list that is intended to help you find missing files:
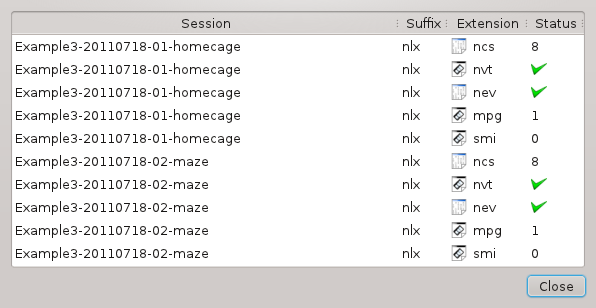
Had a file been missing, e.g. the video for the sleep session, a red cross would have appeared next to its name (see Example 2). Note that ncs files are counted: a missing file would yield a lower count.
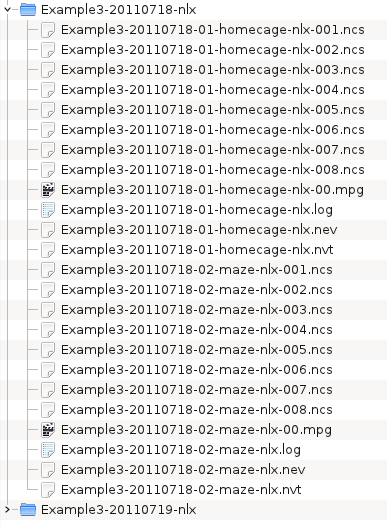
Prepare the other three directories. Your data files should now look like:
Open NDManager. To create a custom template parameter file from a standard template, select -> and select Template-2-tetrodes-32552Hz.xml in /usr/share/ndmanager/templates/. In the General page, enter the name of the experimenter(s) and a description of the experiment (ignore the date and notes). In the Video page, enter the sampling rate (25), width (768) and height (576).
The Anatomical Groups and Spike Groups pages are already filled. Each line lists the channels belonging to the corresponding tetrode: 0 1 2 3 for line 1, and 4 5 6 7 for line 2 (note that channels are numbered from 0). Also indicated are the number of samples per waveform to extract from the wideband data (52 is appropriate for approximately 32 kHz), the location of the peak sample (26, i.e. at the center), and the number of features per channel for the PCA (in practice, a value of 3 is appropriate for a tetrode).

Now go to the Scripts page, click and load all the script descriptions located in /usr/share/ndmanager/descriptions/ (not all of them are relevant here, but you can safely load and ignore them). The scripts are now listed under the Scripts entry. Select them one by one and set the parameter values to suit your needs. Use the Help tabs to get information about the meaning of each parameter. In many cases, default values are provided.
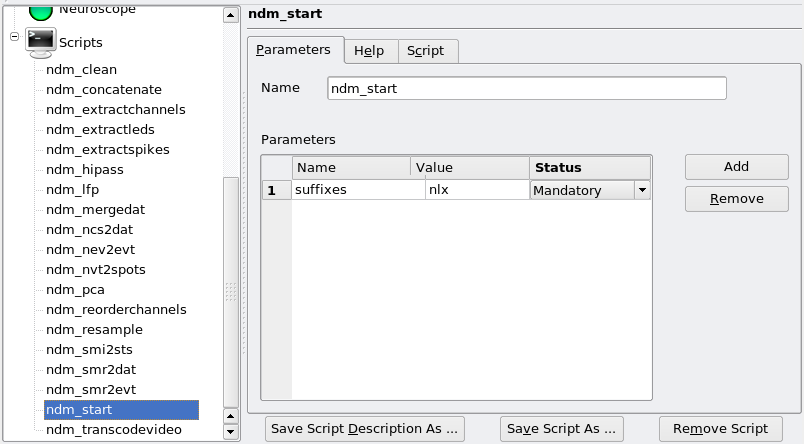
One special (and recurring) parameter deserves special attention: suffixes. This is the suffix that was provided when the data files were prepared, in Recording Setups. In this example, the suffix is nlx. For instance, here is the page for ndm_start:
Save the file as e.g. 2-tetrodes.xml next to your data directories, and quit.
Would you like to make a comment or contribute an update to this page?
Send feedback to the KDE Docs Team