

This page allows you to specify the information needed by NeuroScope. This information is divided into five sections:
Miscellaneous
Video
Clusters
Channel Colors
Channel Offsets
In this page, you can specify the screen gain used to display the traces. In NeuroScope amplitudes are described as follows. By convention, 'x1' corresponds to the current screen gain. Thus, assuming for instance a screen gain of 1 mV/cm, an amplitude of 'x0.38' corresponds to 0.38 mV/cm.
The Background image field allows you to specify an image to be used as a background for the traces. You can use any image format supported by Qt: JPEG, PNG, BMP, XBM, XPM, PNM, MNG and GIF. See your Qt documentation for details (e.g. the QImageIO class description).
In certain behavioral experiments, e.g. when studying place cells or head direction cells, it is necessary to record brain signals as well as the ongoing position and orientation of the animal. NeuroScope has the ability to simultaneously display brain signals and the successive positions of the animal during the same episode. NeuroScope uses the positions extracted from the video recording and stored in a position file.
By default, the positions are drawn against a black background, but an overview of the behavioral maze can also be used as a background image. This image can be flipped and rotated. In addition, an outline of the trajectory of the animal throughout the experiment can be drawn over the background (be it a black background or an image). All this is customizable through the following fields:
Sampling rate (Hz). Default sampling rate of the position file.
Width (pixels). Default horizontal resolution of the video camera.
Height (pixels). Default vertical resolution of the video camera.
Background image. Background image to draw the positions against. You can use any image format supported by Qt: JPEG, PNG, BMP, XBM, XPM, PNM, MNG and GIF. See your Qt documentation for details (e.g. the QImageIO class description).
Draw trajectory. Activate this to draw over the background (be it a black background or an image) an outline of the trajectory of the animal throughout the experiment.
Rotate CCW (degrees). This allows you to rotate (counterclockwise) both the position data and the background image.
flip. This allows you to flip both the position data and the background image.
NeuroScope allows you to browse identified unit activity together with local field potentials. In order to be able to do this, NeuroScope needs the information provided in this page together with the timestamp and the cluster Id of each spike (which are stored in a cluster file and a spike time file respectively).
The Number of samples per waveform and the index of the peak sample allows NeuroScope to highlight the spike waveforms on the traces.
This tab is available only in the Expert Mode.
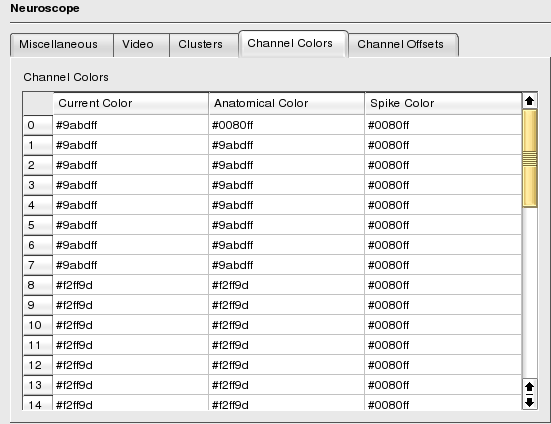
In order to make individual channels as well as channel groups more readily visible, channels can be colored using three different schemes:
By Anatomical Group: A color can be assigned to all the channels in a given anatomical group, this color is referred to as the Anatomical Group color for the given channels.
By Spike Group: Similarly, a color can be assigned to all the channels in a given spike group, this color is referred to as the Spike Group colors for the given channels.
By Channel: Additionally, an individual channel can also be assigned a color.
To change a color, you can either manually type in the html color code or do it graphically. Middle-click on the cell containing the color to modify, this brings up the Color Chooser, as soon as you validate your new choice, the cell value is updated.
Usually the colors are set graphically with NeuroScope, this is why this page is not part of the default mode.
This tab is available only in the Expert Mode.
When the physical organization of your electrodes is more complex than vertical arrays (e.g. epidural electrodes covering the surface of the brain), you might want to store your electrode layout. This can be done by redefining the default offsets. The table contained in this page lists the default offsets.
Usually the default offsets are set graphically with NeuroScope, this is why this page is not part of the default mode.
Would you like to make a comment or contribute an update to this page?
Send feedback to the KDE Docs Team