

This page is available only in the Expert Mode.
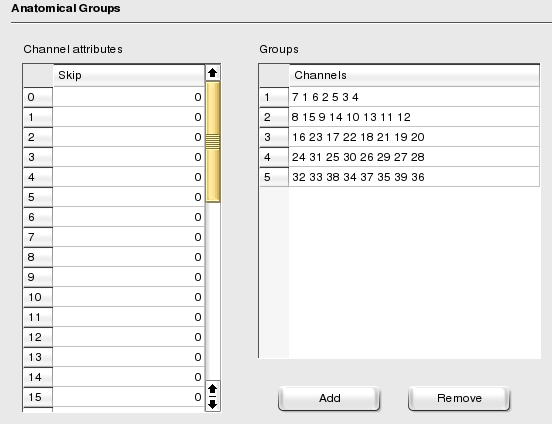
Anatomical groups allow you to classify you channels. Suppose you record data from both the neocortex and the hippocampus: you may want to assign all channels recorded from the neocortex into one group, and all channels recorded from the hippocampus into another group, or maybe have two distinct groups for CA1 and CA3, respectively. This is what anatomical groups are for. More generally however, anatomical groups can be used to implement any classification you may wish, e.g. sites from the same shank in recordings using silicon probes. Usually those groups are created graphically with NeuroScope within the Anatomical Groups Palette (this is why this page is not part of the default mode).
To add a group, press the button; in the newly created cell type in the channel ids separated by a whitespace. To remove groups, select the corresponding rows and press the button. The unlisted channels are not part of any group.
The Channel Attributes table lists the attributes which can be associated to channels. There is currently only one defined attribute, the skip attribute which value can be 0 or 1 (the default value being 0). See the NeuroScope's user manual (section Skipping Channels) for a use of this attribute.
Would you like to make a comment or contribute an update to this page?
Send feedback to the KDE Docs Team